Basic usage#
Installation#
C++#
The installation of the C++ version of Open3D is described under Open3D - C++.
It is required to build it from source.
Python#
Open3D can be installed from master branch with :
PIP :
python -c "import open3d as o3d"``pip install --user --pre https://storage.googleapis.com/open3d-releases-master/python-wheels/open3d-0.11.1-cp38-cp38-linux_x86_64.whl
CONDA :
conda install -c open3d-admin open3d
To quickly test if Open3D is correctly installed, run :
python -c "import open3d as o3d"
You should get a result like <module 'open3d' from 'D:\\Anaconda\\lib\\site-packages\\open3d\\__init__.py'>.
Usage (Python)#
A lot of tutorials are presented in the official documentation.
However, the following snippet has been made to have a complete pipeline as in the PCL Toolkit - Pipeline idea for a quick beginning with Open3D.
It is presented step by step, coded like in a Jupyter Notebook.
First block is used to include necessary imports :
1#Ensure o3d is running
2import open3d as o3d # conda install -c open3d-admin open3d
3import numpy as np
4import time
5from tkinter import Tk #Filepicker
6from tkinter.filedialog import askopenfilename
7Tk().withdraw()
8import copy
9import matplotlib.pyplot as plt
10print(o3d)
Next, we load a file thanks to a file picker. It has a tendency to appear under already opened windows, so watch for it in your application tab.
It uses the method
o3d.io.read_point_cloud(filename) to load a cloud into pcd variable :1#Load pcd file
2print("Load PCD file")
3filename = askopenfilename()
4start_time = time.time()
5pcd = o3d.io.read_point_cloud(filename)
6elapsed_time = time.time() - start_time
7print("Load done in", elapsed_time*1000, "[ms] with", len(pcd.points), "points")
To further apply algorithms, it is often needed to downsample the cloud to have fast running algorithms.
Here, the method is a voxel downsampling, as presented in PCL Toolkit - Downsampling, from previous pcd file
pcd.voxel_down_sample(voxel_size=voxelsize) :1#Downsample it
2voxelsize = 0.8
3print("Downsample the point cloud with a voxel of",voxelsize)
4start_time = time.time()
5downpcd = pcd.voxel_down_sample(voxel_size=voxelsize)
6elapsed_time = time.time() - start_time
7print("Downsample done in", elapsed_time*1000, "[ms] from",len(pcd.points),"to",len(downpcd.points),"points")
To trial the alignment process, a second cloud - moved in space - is created.
Here, we translate it thanks to
moved_down_pcd.translate((xoff,yoff,zoff)) : 1#Creates a moved cloud
2xoff = 33
3yoff = 36
4zoff = 24
5moved_down_pcd = copy.deepcopy(downpcd)
6print("Move cloud with xoff =",xoff,"yoff =",yoff,"zoff =",zoff)
7start_time = time.time()
8moved_down_pcd.translate((xoff,yoff,zoff))
9elapsed_time = time.time() - start_time
10print("Moved cloud done in", elapsed_time*1000)
We can show both clouds in space with :
1#Show
2print("Show cloud - press H to display commands here")
3o3d.visualization.draw_geometries([downpcd,moved_down_pcd])
Giving the following result :
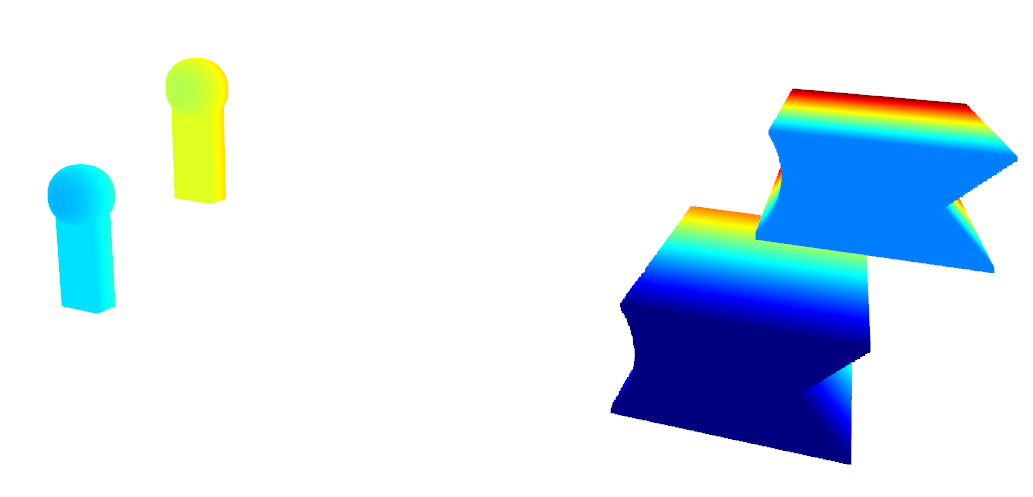
Visualization of the two clouds#
Then, we need to calculate the normals to be able to compute FPFH features :
1#Calculate features for global registration
2 # Add normals
3radius_normal = voxelsize * 2
4print("Estimate normals of base cloud with radius of",radius_normal)
5start_time = time.time()
6downpcd.estimate_normals(o3d.geometry.KDTreeSearchParamHybrid(radius=radius_normal, max_nn=30))
7elapsed_time = time.time() - start_time
8print("Calculated normals in", elapsed_time*1000)
9
10print("Estimate normals of base cloud with radius of",radius_normal)
11start_time = time.time()
12moved_down_pcd.estimate_normals(o3d.geometry.KDTreeSearchParamHybrid(radius=radius_normal, max_nn=30))
13elapsed_time = time.time() - start_time
14print("Calculated normals in", elapsed_time*1000)
15
16
17 # Calculate FPFH
18radius_feature = voxelsize * 5
19print("Compute FPFH feature with search radius %.3f." % radius_feature)
20start_time = time.time()
21pcd_fpfh = o3d.pipelines.registration.compute_fpfh_feature(downpcd,o3d.geometry.KDTreeSearchParamHybrid(radius=radius_feature, max_nn=100))
22elapsed_time = time.time() - start_time
23print("Calculated FPFH in", elapsed_time*1000)
24
25print("Compute FPFH feature with search radius %.3f." % radius_feature)
26start_time = time.time()
27moved_pcd_fpfh = o3d.pipelines.registration.compute_fpfh_feature(moved_down_pcd,o3d.geometry.KDTreeSearchParamHybrid(radius=radius_feature, max_nn=100))
28elapsed_time = time.time() - start_time
29print("Calculated FPFH in", elapsed_time*1000)
Once those data are calculated, we can compute the rough transformation and apply it to the cloud, while visualizing it :
1# Perform global registration
2distance_threshold = voxelsize * 0.5
3print("Calculate global registration with distance threshold %.3f" % distance_threshold)
4start = time.time()
5globreg_result = o3d.pipelines.registration.registration_fast_based_on_feature_matching(
6 moved_down_pcd, downpcd, moved_pcd_fpfh, pcd_fpfh,
7 o3d.pipelines.registration.FastGlobalRegistrationOption(maximum_correspondence_distance=distance_threshold))
8print("Fast global registration took %.3f sec.\n" % (time.time() - start))
9
10moved_down_pcd.transform(globreg_result.transformation)
11o3d.visualization.draw_geometries([downpcd,moved_down_pcd])

Roughly registered clouds#
The clouds can finally be fine aligned thanks to ICP :
1# Perform ICP
2distance_threshold = voxelsize * 0.4
3finereg_result = o3d.pipelines.registration.registration_icp(moved_down_pcd, downpcd, distance_threshold)
4
5moved_down_pcd.transform(finereg_result.transformation)
6o3d.visualization.draw_geometries([downpcd,moved_down_pcd])
And you are done with the transformation piepline !
Clustering#
You can also isolate parts of a cloud thanks to density-based clustering (here, we color each point following their cluster) :
1#Cluster different elements by DBSCAN (density based clustering)
2print("Clustering cloud")
3start_time = time.time()
4with o3d.utility.VerbosityContextManager(o3d.utility.VerbosityLevel.Debug) as cm:
5 clusters = downpcd.cluster_dbscan(eps=6, min_points=40, print_progress=True)
6 labels = np.array(clusters)
7 # eps = distance to neighbors in same cluster
8 # min_points = minimal points in cluster to be considered as is
9elapsed_time = time.time() - start_time
10max_label = labels.max()
11print(f"Point cloud has {max_label + 1} clusters | Done in {elapsed_time*1000} [ms]")
12display(labels)
13
14colors = plt.get_cmap("rainbow")(labels / (max_label if max_label > 0 else 1))
15colors[labels < 0] = 0
16pcd.colors = o3d.utility.Vector3dVector(colors[:, :3])
17o3d.visualization.draw_geometries([pcd])

Clustered parts (each color is a cluster)#